Everything you need to know about stem cells
What are stem cells, and what makes them so unique? What are the different types of stem cells, and how have they impacted modern medical science? Here is us answering all your questions about stem cells.

What if one day we could specifically edit out deadly mutations in the human body? What if we could make targeted edits in the DNA of crops and render them pest resistant, or create gene drives to make infectious vectors like mosquitoes incapable of spreading dangerous diseases? Whether these questions get you thrilled or sceptical, the truth of the matter is that we now have the technology to make all this a reality. Two scientists, Jennifer Doudna and Emmanuelle Charpentier were recently (2020) honoured with the Nobel Prize in chemistry for the discovery of the CRISPR-Cas9 gene-editing technology, a technology that has indeed revolutionised the way we envision biomedical science.
But what exactly is CRISPR-Cas? How has this been used as a tool to edit genomic sequences? What impact does this technology have on our lives and the lives of the generations to come? Let’s find out.
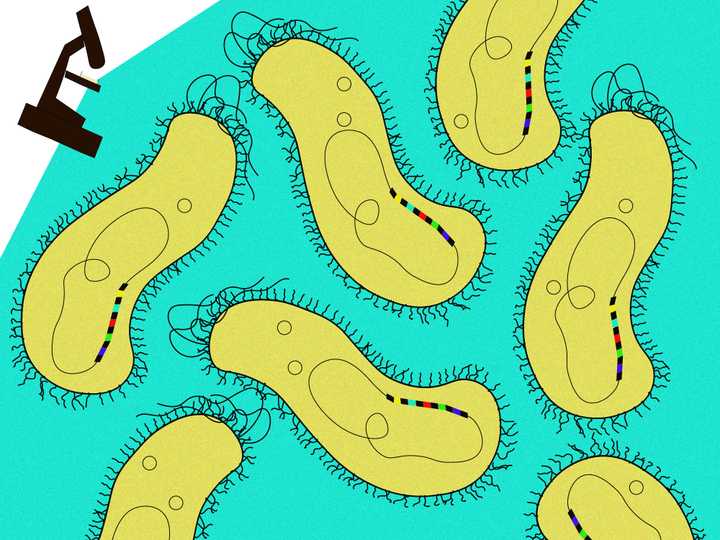
CRISPR (black) and spacer (coloured) sequences were observed in the DNA of many bacteria (yellow). © Sunaina Rao.
Many path-breaking discoveries in science have all begun with a serendipitous observation made by an enthusiastic scientist. The story of CRISPR also began with one such observation. But before we dive into the nitty-gritty of this interesting observation, we need to understand a little about the central dogma of molecular biology.
What is the central dogma of molecular biology?
The genetic material in living organisms is most often composed of DNA (Deoxyribonucleic acid) molecules. DNA is essentially a double-stranded chemical string made of units called nucleotides. These are represented by the letters A, T, G and C. The chemical letters in one strand are complementary to the letters on the other. Hence they attract and bind together, similar to how two strips of velcro cling to each other.
Every gene in the DNA molecule has a unique sequence of these nucleotides and codes for a specific protein. It does this via an intermediate molecule called RNA (Ribonucleic acid). This acts as a messenger, carrying the information coded in the DNA, and helps decode it to make a protein. The RNA is also made of the same chemical letters but is complementary to one of the DNA strands.
This sequence of events, from DNA to RNA to a fully functional protein, is called the central dogma in molecular biology, as the entire functioning of a cell depends on these events. However, more and more studies now show that not all of these messenger RNAs code for proteins. Some seem to have very mysterious and fascinating functions. The RNA that we will come across in this CRISPR story, is also one such RNA!
Now back to the story…
In the 1980s, a group of scientists (Ishino et al., 2016) were interested in deciphering the sequence of a gene that codes for a specific enzyme in the bacteria E. coli. While decoding the sequence of this specific gene, the scientists stumbled upon an interesting set of interspersed repeat sequences elsewhere in the E-Coli genome. An interspersed repeat is essentially a specific sequence of nucleotides of a defined length, that repeats itself at regular intervals. The scientists were not able to deduce at the time, the function of these repeats.
This observation however sparked tremendous curiosity in the scientific community. Within no time these interspaced repetitive sequences were found in many other bacteria (Jansen et al., 2003) and bacteria-like organisms (Mojica et al., 2006).
Now, there were a couple of things extremely intriguing about these repeats.
So these repeats eventually came to be called CRISPR (Clustered Regularly Interspaced Palindromic Repeats; We won’t dwell into what a palindrome is here, but if you are interested, you can check out this article).
This observation got many scientists thinking. Something so commonly found and conserved amongst so many species, must have some function. What did the CRISPR sequences do? This question began another wave of scientific exploration.
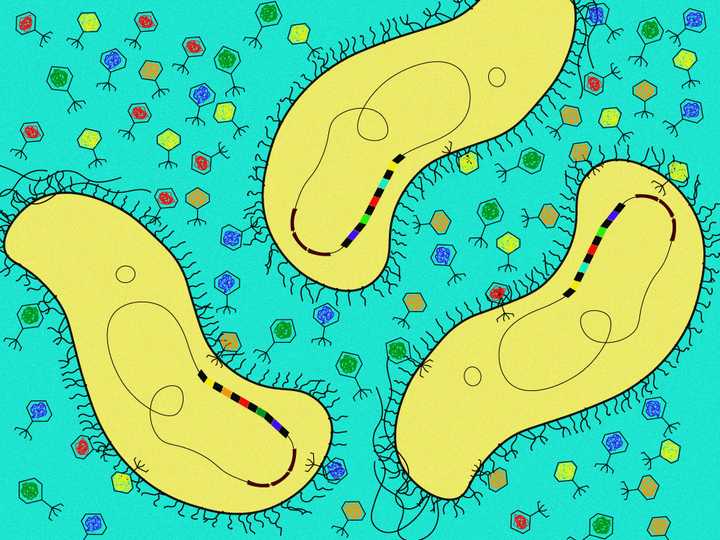
The spacer sequences were largely obtained from bacteriophages (viruses that infect bacteria). The CRISPR sequences were always seen to be accompanied by genes that code for Cas proteins (sequences in brown). © Sunaina Rao.
In the decade to follow, with the arrival of computational techniques, a genomic quest began. As a result of this, a very intriguing reality came to light. The spacer sequences in between the CRISPR repeats did not belong to the bacteria at all. They were alien genes, largely stolen from bacteriophages (viruses that infect bacteria). Not just that, these sequences seemed to provide resistance against the viruses they were taken from (Mojica et al., 2005, Bolotin et al., 2005, Pourcel et al., 2005).
What was even more interesting was that the CRISPR sequences were always associated with another set of genes, which was termed the CRISPR associated genes or Cas genes. These Cas genes were observed to code for Cas proteins that had nucleic acid cleavage function. Meaning they could snip out DNA sequences.
Evidently, these studies brought out the possibility that the CRISPR sequences might be involved in providing immunity to the bacteria against infectious agents such as bacteriophages, and that the mechanism must involve some form of genetic degradation. However, the exact mechanism was still unclear.
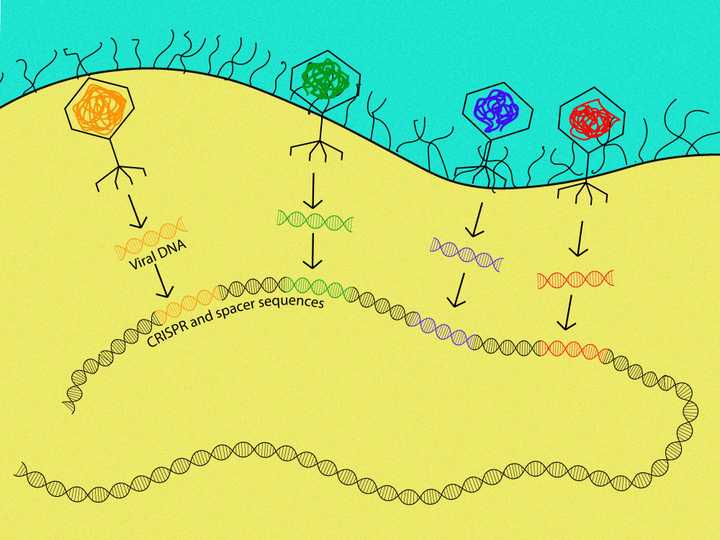
The bacteria stores pieces of viral DNA (coloured sequences) in its own DNA (black). © Sunaina Rao.
In the coming years (the early 2000s), different groups (Horvath and Barrangou, 2010) of scientists (Wiedenheft et al., 2012) began to unravel this mechanism. What became evident was that every time these bacteria were infected by a virus, the bacteria would store a piece of the viral DNA in their own genome, specifically between the CRISPR sequences. It appeared to be some sort of track record of infections.
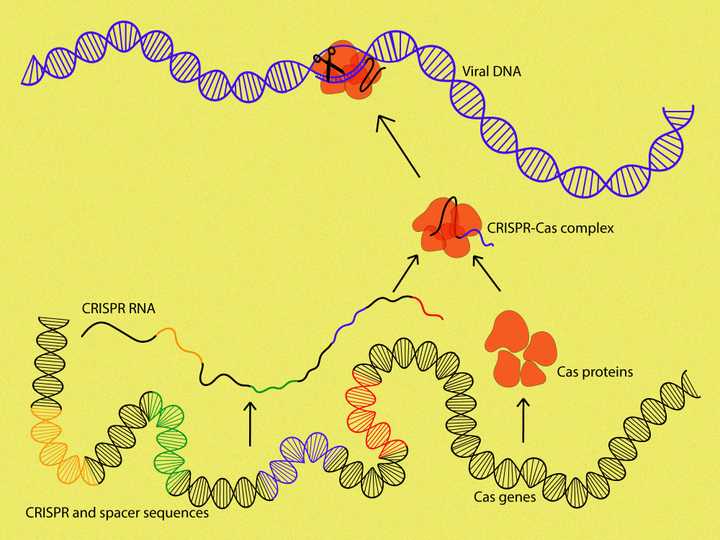
The CRISPR and spacer sequences are transcribed into a CRISPR RNA/crRNA. The crRNA then guides the Cas proteins (orange) to the exact location of the viral DNA (blue), where they bring about cleavage. © Sunaina Rao.
Now, in the case of reinfection by the same virus, the bacterial cell would transcribe the CRISPR sequences along with the adjoining spacer/viral sequences in the DNA into an RNA molecule. This RNA, called CRISPR RNA or crRNA, was complementary to the DNA sequence (Remember what an RNA is?). The spacer sequence in the RNA that correlates with the DNA of the current infecting virus, along with its adjoining CRISPR sequence, would then bind to the Cas proteins to form a complex.
The crRNA, being complementary to the viral DNA sequence, guided the Cas proteins towards the viral genetic material and helped position them in the exact location. The Cas protein, having DNA cutting abilities, would then cut the viral DNA in that specific location.
So essentially the crRNA acted like a GPS tracker accurately finding the invading viral sequence, and the Cas proteins did the dirty job of actually cleaving that sequence. This inevitably led to the elimination of the virus.
Mission accomplished!
But an important question here was why the bacteria didn’t snip its own DNA sequence? After all, the CRISPR RNA was complementary to its own DNA as well, and could surely guide the Cas proteins there. Studies showed that this didn’t happen as the Cas proteins, very intelligently, only cut the DNA sequence that was adjacent to a specific sequence called the PAM sequence (and you guessed it right, this sequence was present only in the viral genome!)
Now isn’t that a phenomenal mechanism of self defense?
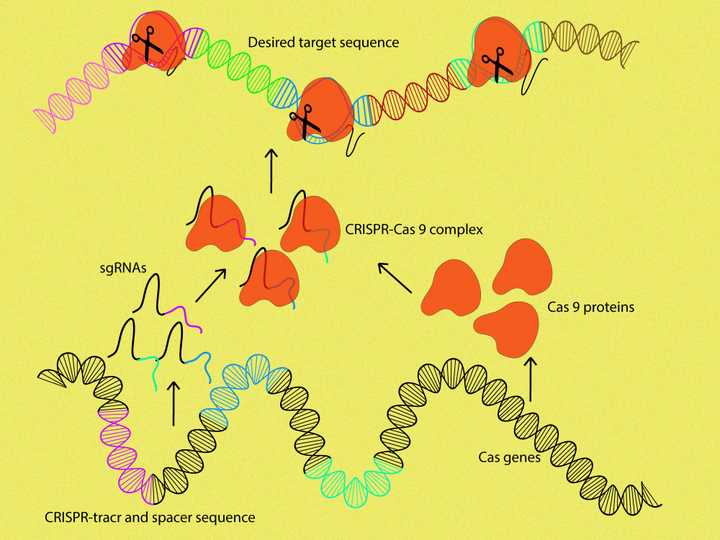
The CRISPR-tracr and spacer sequences are transcribed into corresponding sgRNAs. The sgRNA then guides the Cas9 protein (orange) to the exact location on our desired target sequence (multicoloured sequence on the top) and brings about cleavage. © Sunaina Rao.
The unraveling of the mechanism by which CRISPR sequences function, was by all means an exciting find. It not only opened our eyes to how bacteria defend themselves by cutting up viral DNA, but also made us wonder if we could somehow use the same principle to cut up DNA sequences of our choice.
In order to explore this, a group of scientists led by Jennifer Doudna and Emmanuelle Charpentier (Jinek et al., 2012), began to work on a slightly different type of CRISPR-Cas system. They found that in this system the activity of the Cas protein (in this case called Cas9), required two RNAs, the already known crRNA, and a second RNA called the tracrRNA. They found that the Cas9 would specifically snip the target sequence (similar to what we saw before) on being guided to the exact location by this RNA pair.
However what was more exciting was that they engineered a single RNA out of the two RNAs, called the single guide RNA or sgRNA which could efficiently bind to Cas 9. This engineered RNA could be used to target and snip out any sequence of our choice. On being snipped, the cell would try to repair it, introducing mutations in the process. These mutations could now render the gene inactive.
It is not surprising that scientists soon extended this technology (Mali et al., 2013) to edit not just one, but multiple genes (Cong et al., 2013), in the human genome.
Imagine for a second, if we could edit any kind of DNA sequence, exactly where we wanted, what all could come out of it?
This technology indeed opened doors to immense possibilities (Hsu et al., 2014). Thanks to the CRISPR technology, we could now engineer cells and animals in the lab that have very specific cuts and mutations in their genome, thereby making it easy to study the function of those genes. This tool is now being advanced in so many ways that in the future, we might also be able to specifically replace faulty genes with healthy ones in individuals with genetic diseases such as sickle cell anaemia, thalassemia, muscular dystrophy or certain types of cancer such as melanoma. We could create mosquitoes that no longer have the ability to spread diseases such as malaria. We might also be able to create crops that are resistant to specific diseases.
Never before in the history of biomedicine could we get this creative. Considering the fact that we have the entire sequence information of the human genome, we could modify genes at will. We could literally design our offspring to be fitter, more intelligent, more good looking and so on. Now, as exciting as this may sound, it is also that scary.
Is it ethically right to genetically design your own offspring and thereby reshape the course of the coming generations?
Also, is it scientifically advisable? It is true that we now know a lot about the functioning of the human body. But there is still a lot we don’t know. Making inheritable changes in the human genome, might bring massive setbacks in human evolution as a whole. Hence, strict regulations in the use of this technology in the human population is deemed mandatory, not just by the creators of the technology but by the scientific community as a whole.
What’s most fascinating however is that a simple observation made by a group of scientists, more than two decades ago, led to the discovery of a tool that could impact mankind in ways unimaginable. Hopefully one day we will use this tool to positively transform medical science, while still respecting the scientific and ethical boundaries.